PRP021: Data on Patient Record Trajectory for Linkage (DataPRinT Linkage)
Aashka Bhatt, BSc, MSc; Tao Chen; Conrad Pow; Rahim Moineddin, PhD; Babak Aliarzadeh, MD, MPH; Steven Bernard; Michelle Greiver, MD, MSc, CCFP
Abstract
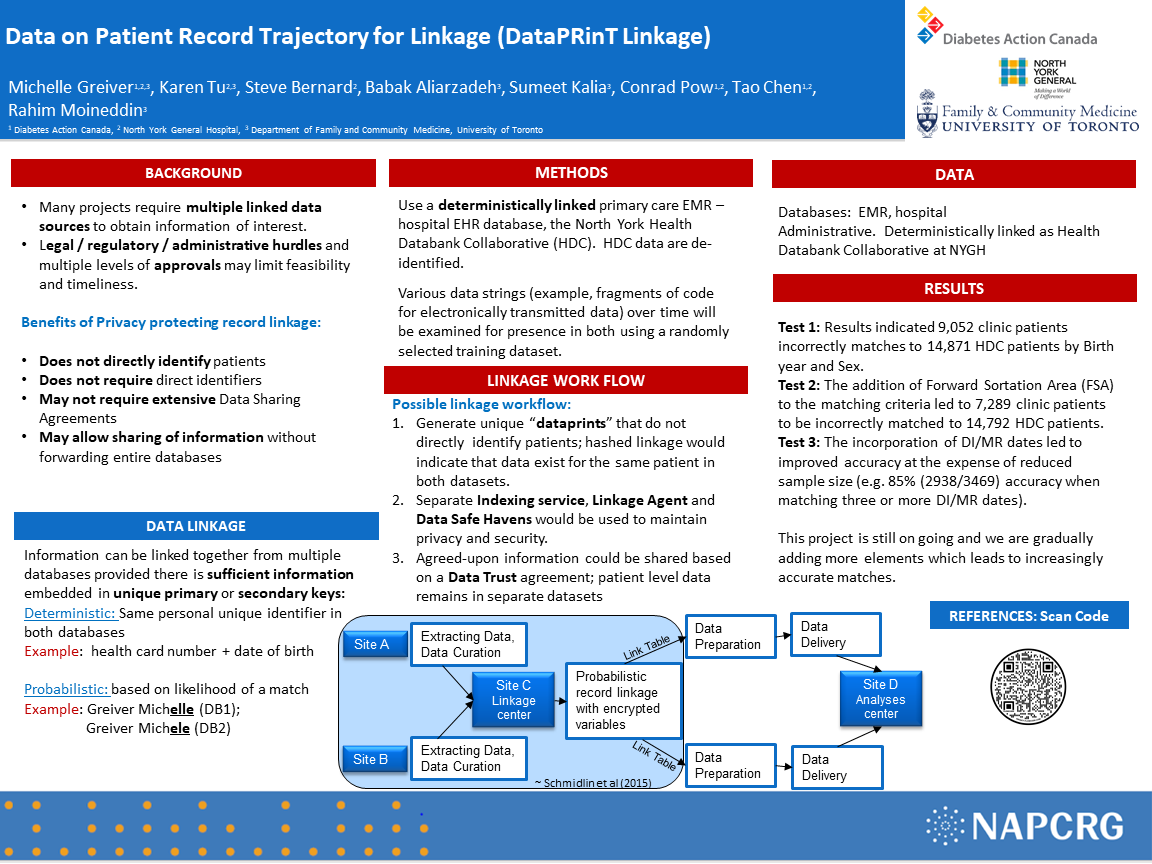
BACKGROUND: The linkage of Electronic Medical Records (EMR) with other data sources is highly valuable for research and health system monitoring. Once linked, combined resources can be analyzed to provide answers to a variety of health questions.
STUDY DESIGN: Data strings included birth year, sex, first three letters of postal code, diagnostic imaging or medical report (DI/MR). These identifiers do not directly identify patients and therefore used as strings in the selected Dataprints, as unique linkage variables.
DATASET: Health Databank Collaborative (HDC), University of Toronto Practice-Based Research Network) and North York General Hospital databases.
OBJECTIVES: To develop a method to probabilistically link the patient’s health trajectory using primary care EMR data with administrative data, without the need to transfer large datasets or identifiable information.
OUTCOME MEASURES: Linkage quality will be assessed by the number of true matches and represented by sensitivity, specificity, positive and negative predictive values.
RESULTS: Results indicated 9,052 clinic patients incorrectly matches to 414,871 HDC patients by Birth year and Sex. The addition of Forward Sortation Area (FSA) to the matching criteria led to 7,289 clinic patients to be incorrectly matched to 14,792 HDC patients. The incorporation of DI/MR dates led to improved accuracy at the expense of reduced sample size (e.g. 85% (2938/3469) accuracy when matching three or more DI/MR dates).
CONCLUSIONS: Preliminary results informed processes to enable analyses across datasets while adhering to privacy legislation.
STUDY DESIGN: Data strings included birth year, sex, first three letters of postal code, diagnostic imaging or medical report (DI/MR). These identifiers do not directly identify patients and therefore used as strings in the selected Dataprints, as unique linkage variables.
DATASET: Health Databank Collaborative (HDC), University of Toronto Practice-Based Research Network) and North York General Hospital databases.
OBJECTIVES: To develop a method to probabilistically link the patient’s health trajectory using primary care EMR data with administrative data, without the need to transfer large datasets or identifiable information.
OUTCOME MEASURES: Linkage quality will be assessed by the number of true matches and represented by sensitivity, specificity, positive and negative predictive values.
RESULTS: Results indicated 9,052 clinic patients incorrectly matches to 414,871 HDC patients by Birth year and Sex. The addition of Forward Sortation Area (FSA) to the matching criteria led to 7,289 clinic patients to be incorrectly matched to 14,792 HDC patients. The incorporation of DI/MR dates led to improved accuracy at the expense of reduced sample size (e.g. 85% (2938/3469) accuracy when matching three or more DI/MR dates).
CONCLUSIONS: Preliminary results informed processes to enable analyses across datasets while adhering to privacy legislation.
Jack Westfall
jwestfall@aafp.org 11/21/2021Great poster and abstract. looking forward to the next iteration of results. NAPCRG 2022! Thanks for sharing at NAPCRG